What else is out there?
[HAL 2.1] To What Extent Current Limits of Phylogenomics Can Be Overcome?
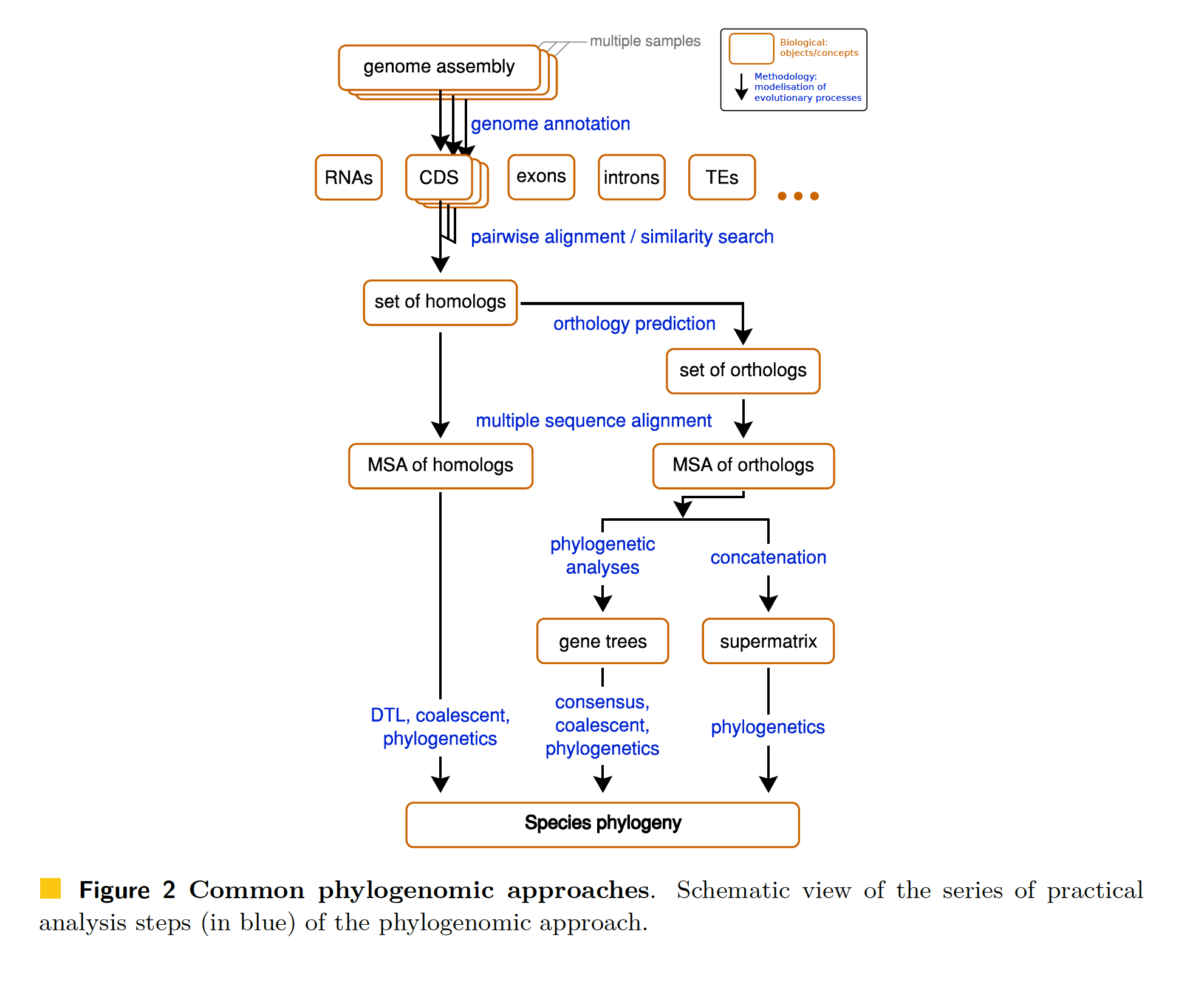
Concept flowchart
Phylogenomic pipeline (HAL 2.1)
What do we know now?
- Motivation: phylogenomics pipeline
- Intro Illumina, quality control, assembly, alignment, filtering
- Alignment
- Reading: HAL 2.2, ClustalW, MUSCLE, T-coffee papers
- Software: T-coffee, Clustal, Muscle
- Orthology detection
- Reading: HAL 2.4, Nichio 2017
- Software: Many options in class google slides!
- Overview of phylogenetic inference: criteria score, search in tree space
- Gene tree estimation
- Distance methods
- Reading (supplemental): HB Ch 5, Baum Ch 8
- Software: R packages ape, phangorn
- Parsimony methods
- Reading (supplemental): HB Ch 8, Baum Ch 7
- Software: R packages ape, phangorn
- Models of evolution
- Maximum likelihood
- Reading: HAL 1.2, RAxML and IQ-Tree papers
- Software: RAxML (HAL 1.3) and IQ-Tree
- Bayesian
- Reading: HAL 1.4, Mascimento 2017, MrBayes papers
- Software: MrBayes (HB Ch 7)
- Model selection (invited guest: Rob Lanfear developer of IQ-Tree)
- Species tree/network estimation: the coalescent model
- Reading: HAL 3.1, 3.3, ASTRAL and BUCKy papers
- Software: PhyloNetworks wiki pipeline (MDL, RAxML, MrBayes, BUCKy, ASTRAL, SNaQ)
- Co-estimation methods
- Reading: BEAST papers
- Taming the BEAST slides
Main conclusions
- Phylogenomics is hard
- Importance of data quality and phylogenetic signal
- Importance of clear description of methods used, assumptions and limitations
- Make sure that you know (in general) what the method you are using does
- Importance of model selection and model fit
- Importance of measures of confidence
- No desperate quest for 100 bootstrap support values!
- Importance of reproducibility
- Every choice matters, so keep good track of the choices
Good news
- This was not the class to make you all experts in phylogenetics
- Class notes are publicly available on github forever
- Upcoming: lecture YouTube videos publicly available (Spring 2024)
- You have now your own notes in your personal github repository
- You can continue to have support from phylogenetics student community in slack (if you choose to remain in the workspace)
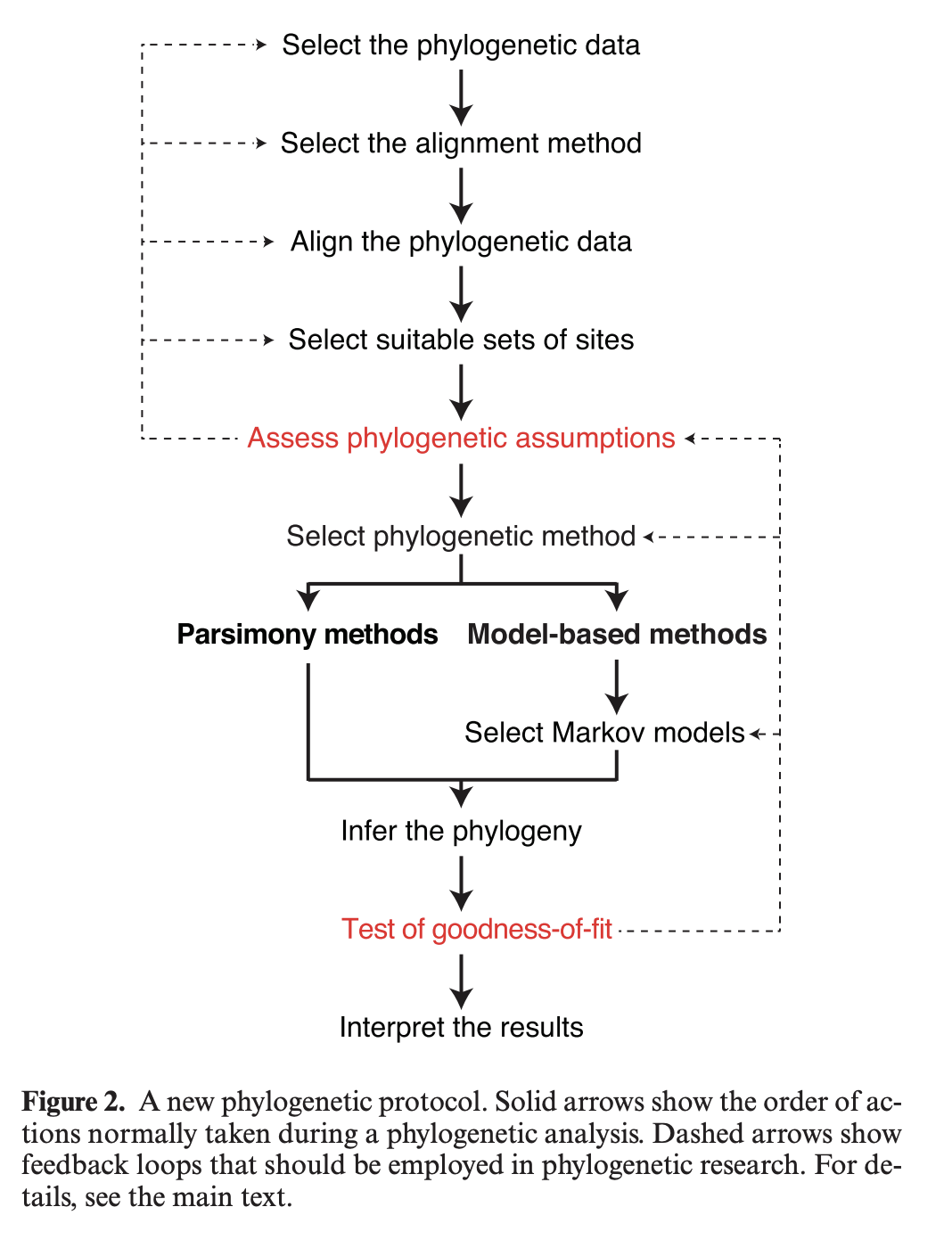
Jermiin et al 2020
What else is out there?
- Dating
- Reading: HAL 5.1
- Supplemental reading: HB Ch 11
- Bayesian dating: Bromham 2017
- Software
- Reconciliation
- Species delimitation
- Reading: HAL 5.5
- Software:
- BPP: HAL 5.6
- iBPP (morphology and genes): https://github.com/cecileane/iBPP
- SNP methods
Final housekeeping
- Project deadline: May 7th
- Presentations: May 1 and May 6
- 6-minute presentations (very strict!)
- Please send me your slides at 12pm (or earlier) on the day you are presenting
- Official student evaluations
- Forward me the email by May 5th to receive HW credit
- Class feedback google form (link on canvas and slack)
- Highly appreciated
- Less than 5 minutes to respond
- Advice for future students: slides